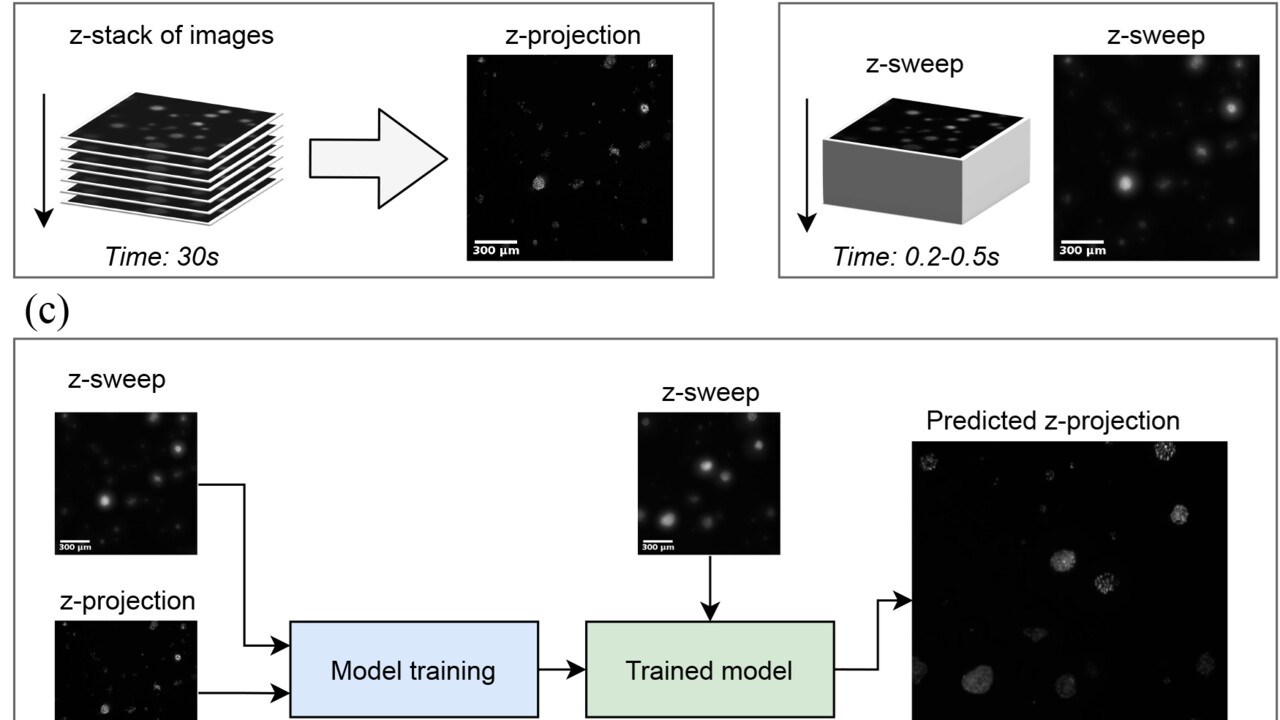
Information about the scientific article:
Forsgren E, Edlund C, Oliver M, Barnes K, Sjögren R, Jackson TR (2022): High-throughput widefield fluorescence imaging of 3D samples using deep learning for 2D projection image restoration. PLoS ONE 17(5): e0264241.