
Image: Mattias Pettersson

Image: Mattias Pettersson
Research group Our laboratory investigates the molecular mechanisms governing autophagy (recycling process in cells) and membrane trafficking (intracellular transport) through the development and application of innovative chemical and chemo optogenetic methodologies.
Yaowen Wu Lab concept
ImageYaowen WuLiving systems rely on precisely regulated chemical reactions that occur with spatial and temporal specificity in response to extracellular or intracellular cues. To decipher these processes, one must both visualize and perturb the underlying chemistry. Chemical tools thus offer unparalleled capabilities to emulate or modulate biological pathways, facilitating both mechanistic investigations and the engineering of novel cellular functions (synthetic biology).
We pursue two complementary research strands:
Together, these integrated approaches enable the discovery of previously unrecognized regulatory mechanisms and the creation of new functional modalities.
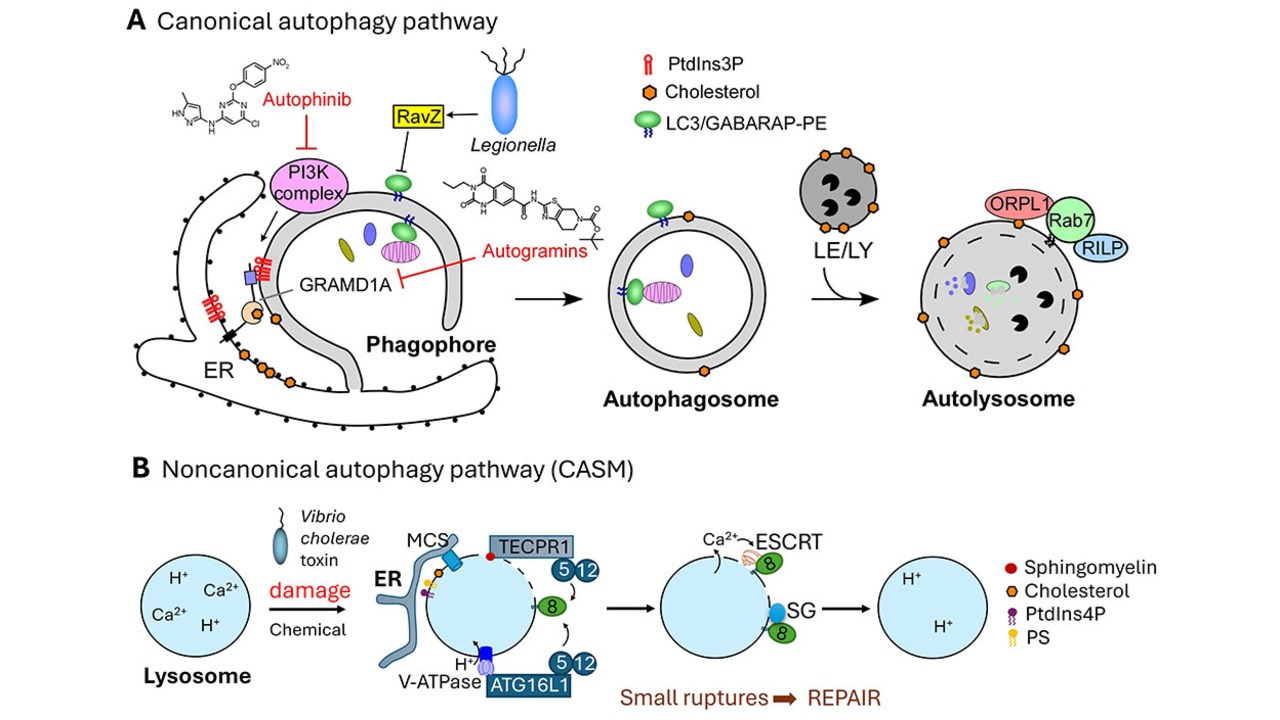
Autophagy is an evolutionarily conserved catabolic pathway in eukaryotic cells, responsible for the clearance of damaged organelles and aggregated proteins via sequestration within double‑membraned autophagosomes. Dysregulation of autophagy contributes to a spectrum of pathologies—including cancer, neurodegeneration, and infectious disease.
ImageYaowen Wu
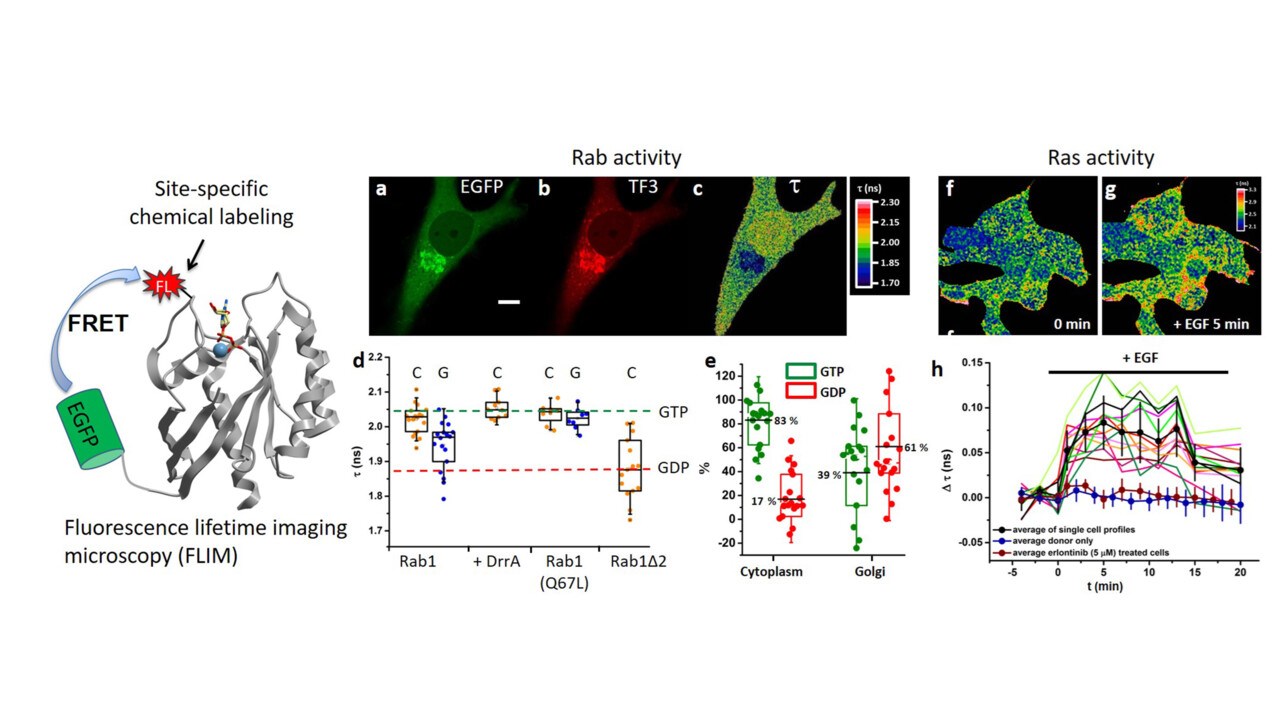
Eukaryotic cells maintain compartmentalization through a sophisticated vesicular transport network. Rab GTPases act as master regulators of vesicle formation, targeting, and fusion by cycling between active and inactive states under the control of specific regulators and effectors. We investigate how these elaborate reactions are integrated at both molecular and cellular scales and how their dysregulation leads to disease. Our quantitative analyses of Rab GTPase membrane targeting have produced a comprehensive model for their localization—a framework that illuminates general principles of intracellular transport (PNAS 2014, PNAS 2016, Biochemistry 2019, MiMB 2021, MiMB 2021).
ImageYaowen Wu
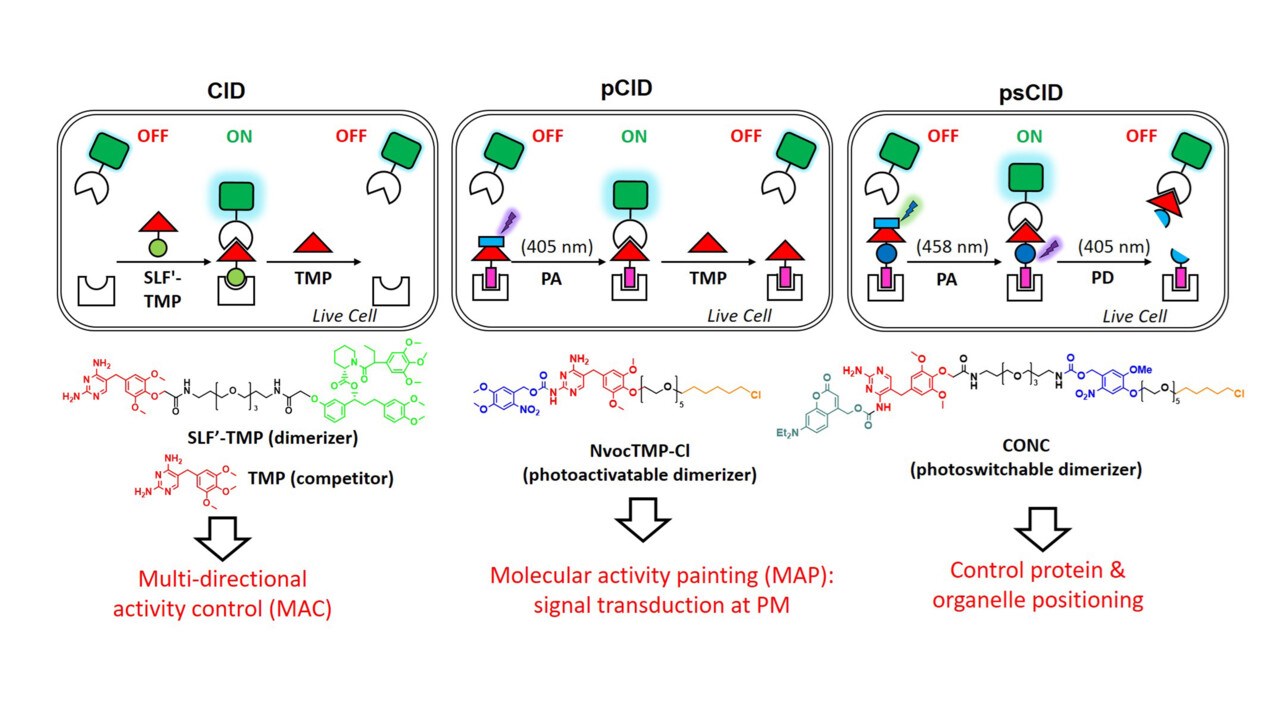
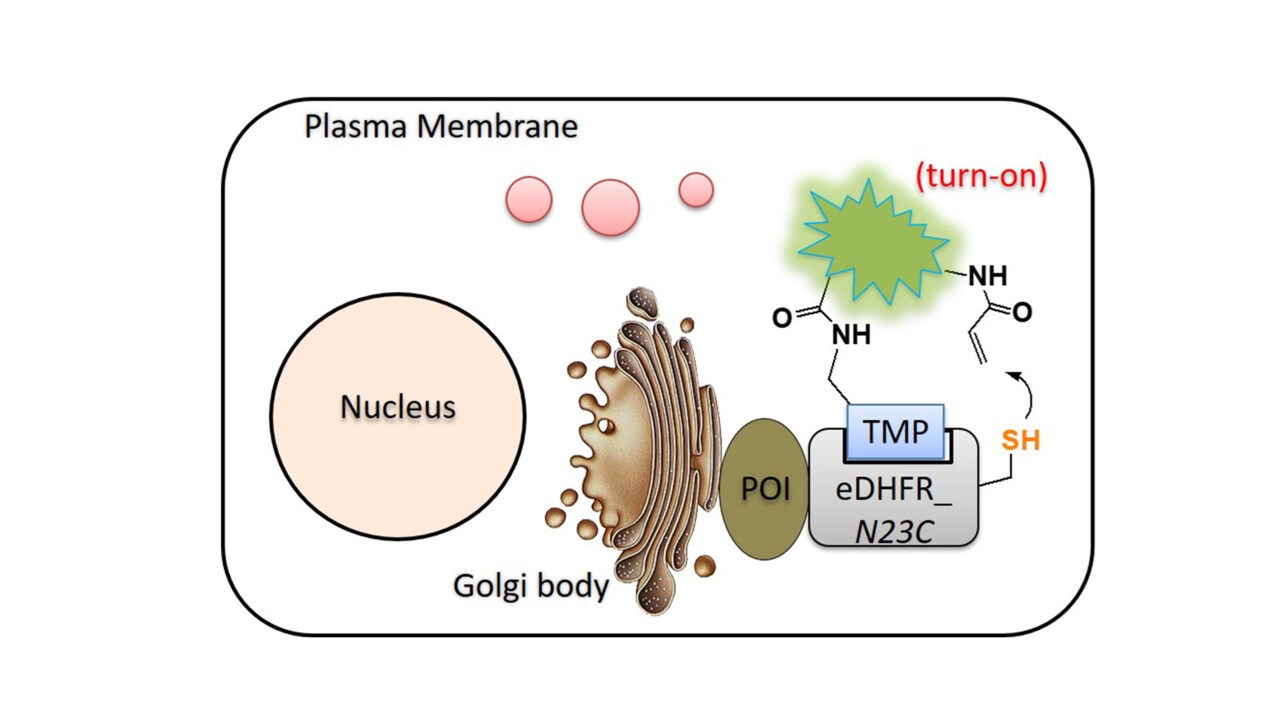
Genetic perturbations (e.g., overexpression, knockout, knockdown) provide important insights but operate on slow timescales (hours to days), often obscuring transient phenotypes. In contrast, chemical and light‑inducible dimerization systems afford unmatched spatiotemporal precision for controlling protein activity. They have been very useful to dissect the complex biological mechanisms (JACS 2012, Curr Opin Chem Biol 2015, ChemEurJ 2019, Nat Meth 2023, ChemPhotoChem 2026, reviews).
ImageYaowen Wu
The installation of synthetic probes—such as fluorophores, affinity tags, and specialized labels—onto proteins is essential for functional characterization in vitro and in vivo. Chemical methods have significantly broadened the toolkit for protein modification.
These chemical biology innovations not only drive our own research but also contribute broadly applicable tools to the life science community.
ImageYaowen Wu
Yaowen Wu received his BS in Chemistry from Sun Yat-sen University in 2001 and his MS in Organic Chemistry from Tsinghua University in 2004 in China. After graduating as Dr rer. nat. (2008) at the Technische Universität Dortmund working at the Max Planck Institute of Molecular Physiology in Germany and a postdoctoral study in cell biology at King’s College London, he has been leader of an Otto Hahn group at the Max Planck Institute in Dortmund since 2010. Since 2012, he has been group leader of Chemical Genomics Centre of the Max Planck Society. He was then appointed as Professor in Biochemistry at the Umeå University in 2018. He has served as Director of Umeå Centre for Microbial Research (UCMR) since 2020.
His research group develops small molecules and new chemical methods to modify proteins or manipulate protein function in the context of biological systems with a particular focus on regulatory mechanisms in membrane trafficking and autophagy. He has received awards and honors including Göran Gustafsson Prize in Molecular Biology “for his innovative molecular studies of intracellular transport and autophagy” (awarded by Royal Swedish Academy of Sciences annually to one outstanding scientist of < 45 years old in each field of mathematics, physics, chemistry, molecular biology and medicine), "Future of Biochemistry" recognized by “tackling problem that transcend traditional field boundaries and challenges” in the field of biochemistry, Wallenberg Academy Fellow (Royal Swedish Academy of Sciences), European Research Council (ERC) Investigator, Behrens-Weise Award, Biomedicine Research Prize and Otto-Hahn Award (selected as the single awardee of the Otto-Hahn Group Leader in 2009 in the biomedical section of the Max Planck Society).
Zhang J, Herzog LK, Corkery DP, Lin TC, Klewer L, Chen X, Xin X, Li Y, Wu YW*. (2025) Modular Photoswitchable Molecular Glues for Chemo-Optogenetic Control of Protein Function in Living Cells. Angew Chem Int Ed. e202416456. (featured as Hot Paper)
Knyazeva A, Li S, Corkery D, Shankar K, Herzog L, Zhang X, Singh B, Niggemeyer G, Grill D, Gilthorpe J, Gaetani M, Carlson LA, Waldmann H, Wu YW*. (2024) A chemical inhibitor of IST1-CHMP1B interaction impairs endosomal recycling and induces noncanonical LC3 lipidation. Proc. Natl. Acad. Sci. U. S. A. 121 (17), e2317680121.
Corkery DP, Castro-Gonzalez S, Knyazeva A, Herzog LK, Wu YW*. (2023) An ATG12-ATG5-TECPR1 E3-like complex regulates unconventional LC3 lipidation at damaged lysosomes. EMBO Rep: e56841.
Comment in Florey O. TECPR1 helps bridge the CASM during lysosome damage. EMBO J. 2023: e115210; Comment in Deretic V, Klionsky, DJ. (2024) An expanding repertoire of E3 ligases in membrane Atg8ylation. Nat Cell Biol. 26: 307–308.
Laraia L, Friese A, Corkery DP, Konstantinidis G, Erwin N, Hofer W, Karatas H, Klewer L, Brockmeyer A, Metz M, Schölermann B, Dwivedi M, Li L, Rios-Munoz P, Köhn M, Winter R, Vetter IR, Ziegler S, Janning P, Wu YW*, Waldmann H*. (2019) The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis. Nat. Chem. Biol. 15(7):710-720.
Comment in Aldrich LN. Lipids lead the way. Nat. Chem. Biol. 2019, 15(7):653-654
Chen X, Wu YW*. (2018) Tunable and photoswitchable chemically induced dimerization for chemo-optogenetic control of protein and organelle positioning. Angew. Chem. Int. Ed. 57 (23): 6796-6799.
Yang A, Pantoom S, Wu YW*. (2017) Elucidation of anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins. eLife. pii: e23905.
Chen X, Venkatachalapathy M, Kamps D, Weigel S, Kumar R, Orlich M, Garrecht R, Hirtz M, Niemeyer CM, Wu YW*, Dehmelt L*. (2017) “Molecular Activity Painting”: Switch-like, light-controlled perturbations inside living cells. Angew. Chem. Int. Ed. 56 (21):5916-5920 (Hot paper, inside cover story)
Voss S, Krüger DM, Koch O, Wu YW*. (2016) Spatiotemporal imaging of small GTPases activity in live cells. Proc. Natl. Acad. Sci. U. S. A. 113 (50):14348-14353.
Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann JE, Theiss C, Dehmelt L, Wu YW*. (2014) A bioorthogonal small-molecule switch system for controlling protein function in live cells. Angew. Chem. Int. Ed. 53 (38):10049-55.
Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu YW*. (2014) The role of the hypervariable C-terminal domain in Rab membrane targeting. Proc. Natl. Acad. Sci. U. S. A. 111 (7): 2572-7. Recommended by Faculty1000

Molecular sensors that detect holes in lysosomal membranes for rapid repair have been identified.

Harshit Malhotra's research aims to use special tools to control the cellular processes of bacteria.

Professor Yaowen Wu's lab is at the forefront of developing chemo-optogenetic systems.